|
Ralph Gauges, Ursula Rost, Sven Sahle and Katja Wegner
European Media Laboratory
Schloss-Wolfsbrunnen Weg 33
69118 Heidelberg
Germany
October 28, 2003
With SBML there now is a common standard for the exchange of dynamical systems data which has already been adopted by many applications in this field [1,2,3]. Since SBML had no means of storing layout information for reaction networks, we developed an extension to SBML that would allow us to store this layout information in SBML files. There already exists an extension to SBML by Herbert M. Sauro that deals with layout information in SBML files [5] well tailored to his program JDesigner [4]. However, in order to provide generality, our specification tries to limit the extensions to just specifying information that concerns the placement of the objects and leaves the rendering to the application.
The overall structure of this proposal reflects some design decisions that will be explained in this paragraph. These decisions are mainly based on the discussion on the mailing list.
First it was requested that it should be possible to have several layouts in one sbml file. This leads to the obvious choice to have a listOfLayouts outside the model part of the sbml file instead of direct annotations to the model elements.
The next question is how tight the relation between the model and the layout should be. It was requested that there should be no strict one-to-one connection between model elements and layout elements. Therefore the layout part of the sbml file cannot just duplicate the structure of the model part. This leads to a structure where a layout contains several lists of layout elements (compartmentGlyphs, speciesGlyphs, ...) There seems to be consensus that one model element can be represented by several layout elements. For example it can be useful to have several representations of one species in the layout to avoid lots of crossing arrows. This can be accomplished if every layout element has a field that refers to the id of a model element.
We also think that there are cases where a layout element does not correspondent to exactly one model element. This could occur if the layout shows a simplified version of the model where one reaction in the layout correspondents to several reactions and intermediate species in the model. This is the reason why the field in the layout elements that refers to the model elements is optional.
Further on we think that the layout should be described in biochemical terms (species, reactions, ...) and not in terms of graph theory (nodes, edges). Otherwise an existing language for graph layouts could be used.
The result of all this is a way to describe a graphical layout of a reaction network in biochemical terms. This layout can be closely tied to the biochemical model. A graphical model editor for example would typically create a layout that is closely connected (by a one-to-several relation from the model elements to the layout elements) to the model. A more general layout design program could also create a layout that is not so closely tied to the model, for example it could create a layout that shows a simplified version of the model.
Last but not least we decided to separate between layout information and render information. By layout information we mean the position and size of all the layout elements and their relations. Render information would be colors, line widths, bitmaps, ... While every program dealing with layouts should be able to read and write the layout information the render information could be ignored or could have program specific extensions. This proposal concentrates on the layout part because we thought it would be easier to reach an agreement on the overall structure of the layout without the rendering details. The rendering detail are described in another document for now, but we expect the documents to be merged eventually.
All size information given for layout objects are understood to be pt, which is defined to be 1/72 of an inch. This will be consistent with the way font sizes will be specified in the render part of the diagram.
For the extensions we use a separate namespace of the following form
xmlns:sl2="http://projects.eml.org/bcb/sbml/level2". A SBML file that would utilize the extension could have the following form:
<?xml version="1.0" encoding="UTF-8"?>
<sbml xmlns:sbml="http://www.sbml.org/sbml/level2" level="2" \\
version="1"
xmlns:sl2="http://projects.eml.org/bcb/sbml/level2"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://projects.eml.org/bcb/sbml/level2
http://projects.eml.org/bcb/sbml/level2/layout2.xsd">
XML Schema representation:
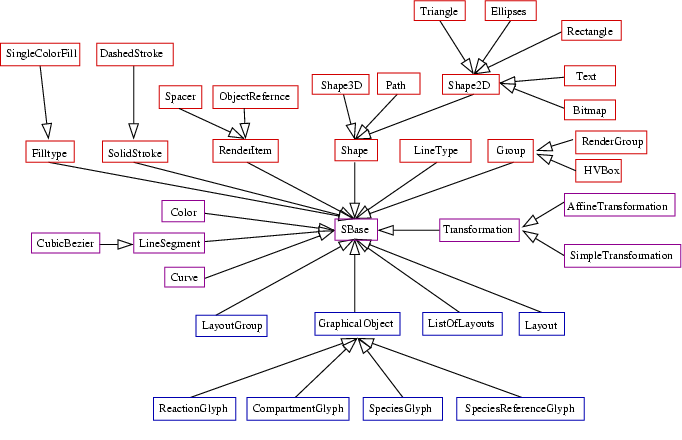
<xsd:complexType name="SBase" abstract="true">
<xsd:sequence>
<xsd:element name="notes" minOccurs="0">
<xsd:complexType>
<xsd:sequence>
<xsd:any namespace="http://www.w3.org/1999/xhtml"
processContents="skip"
maxOccurs="unbounded"/>
</xsd:sequence>
</xsd:complexType>
</xsd:element>
<xsd:element name="annotation" minOccurs="0">
<xsd:complexType>
<xsd:sequence>
<xsd:any processContents="skip" maxOccurs="unbounded"/>
</xsd:sequence>
</xsd:complexType>
</xsd:element>
</xsd:sequence>
<xsd:attribute name="metaid" type="xsd:ID" use="optional"/>
</xsd:complexType>
XML Schema representation:
<xsd:simpleType name="SId">
<xsd:restriction base="xsd:string">
<xsd:pattern value="(_|[a-z]|[A-Z])(_|[a-z]|[A-Z]|[0-9])*"/>
</xsd:restriction>
</xsd:simpleType>
<xsd:complexType name="ListOfCompartmentGlyphs">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="compartmentGlyph" type="sl2:CompartmentGlyph"
maxOccurs="unbounded"/>
</xsd:sequence>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="ListOfSpeciesGlyphs">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="speciesGlyph" type="sl2:SpeciesGlyph" maxOccurs="unbounded"/>
</xsd:sequence>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="ListOfReactionGlyphs">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="reactionGlyph" type="sl2:ReactionGlyph" maxOccurs="unbounded"/>
</xsd:sequence>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="ListOfAdditionalGraphicalObjects">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:any namespace="##other" processContents="skip"
minOccurs="0" maxOccurs="unbounded"/>
</xsd:sequence>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="ListOfGroups">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="group" type="sl2:LayoutGroup" maxOccurs="unbounded"/>
</xsd:sequence>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="Layout">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="listOfCompartmentGlyphs"
type="sl2:ListOfCompartmentGlyphs" minOccurs="0"/>
<xsd:element name="listOfSpeciesGlyphs" type="sl2:ListOfSpeciesGlyphs"
minOccurs="0"/>
<xsd:element name="listOfReactionGlyphs" type="sl2:ListOfReactionGlyphs"
minOccurs="0"/>
<xsd:element name="listOfAdditionalGraphicalObjects"
type="sl2:ListOfAdditionalGraphicalObjects" minOccurs="0"/>
<xsd:element name="listOfGroups" type="sl2:ListOfGroups" minOccurs="0"/>
</xsd:sequence>
<xsd:attribute name="id" type="sl2:SId"/>
<xsd:attribute name="width" type="xsd:double"/>
<xsd:attribute name="height" type="xsd:double"/>
<xsd:attribute name="depth" type="xsd:double" use="optional" default="0.0"/>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="ListOfLayouts">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="layout" type="sl2:Layout" maxOccurs="unbounded"/>
</xsd:sequence>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:element name="listOfLayouts" type="sl2:ListOfLayouts"/>
Right now, our proposal contains two types of transformation which are both derived from an abstract base class called Transformation which in turn is derived from SBase. The Transformation class does not add any new attributes to SBase. From Transformation we derive a class SimpleTransformation which has some attributes to specify translational (tx,ty,tz), rotational (rx,ry,rz) and scaling (sy,sy,sz) transformations for all three axes. All attributes are double values and optional. The default values are 1.0 for the three scaling attributes and 0.0 for the other six attributes. The SimpleTransformation will probably cover 99% of what users need, but in case this is not enough, we have a second type of transformation, the AffineTransformation. The AffineTransformation specifies a 4x3 transformation matrix. The parameters are aligned in rows with 4 parameters each. The parameters are called a0,a1,a2,a3 for row one b0,b1,b2,b3 for row 2 and c0,c1,c2,c3 for row 3. All parameters are optional and the defaults are such that the identity matrix results.
<xsd:complexType>
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="transformation" type="sl2:Transformation" minOccurs="0" maxOccurs="1"/>
</xsd:sequence>
<xsd:attribute name="id" type="sl2:SId"/>
<xsd:attribute name="x" type="xsd:double"/>
<xsd:attribute name="y" type="xsd:double"/>
<xsd:attribute name="z" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="width" type="xsd:double"/>
<xsd:attribute name="height" type="xsd:double"/>
<xsd:attribute name="depth" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="renderObject" type="sl2:SId"/>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType>
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="SimpleTransformation">
<xsd:complexContent>
<xsd:extension base="sl2:Transformation">
<xsd:attribute name="tx" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="ty" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="tz" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="rx" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="ry" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="rz" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="sx" type="xsd:double" use="optional" default="1.0"/>
<xsd:attribute name="sy" type="xsd:double" use="optional" default="1.0"/>
<xsd:attribute name="sz" type="xsd:double" use="optional" default="1.0"/>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="AffineTransformation">
<xsd:complexContent>
<xsd:extension base="sl2:Transformation">
<xsd:attribute name="a0" type="xsd:double" use="optional" default="1.0"/>
<xsd:attribute name="a1" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="a2" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="a3" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="b0" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="b1" type="xsd:double" use="optional" default="1.0"/>
<xsd:attribute name="b2" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="b3" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="c0" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="c1" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="c2" type="xsd:double" use="optional" default="1.0"/>
<xsd:attribute name="c3" type="xsd:double" use="optional" default="0.0"/>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
XML Schema representation:
<xsd:complexType name="CompartmentGlyph"> <xsd:complexContent> <xsd:extension base="sl2:GraphicalObject"> <xsd:attribute name="compartment" type="sl2:SId" use="optional"/> </xsd:extension> </xsd:complexContent> </xsd:complexType>
example:
.
.
.
<compartment id="compartment" volume="1"/>
.
.
.
<sl2:listOfCompartmentGlyphs>
<sl2:compartmentGlyph id="cGlyph" compartment="compartment"
x="10.0" y="10.0" w="60" h="50"/>
</sl2:listOfCompartmentGlyphs>
.
.
.
Since an sbml document can contain species that don't appear in any reaction a species can have zero or more representations on screen which are represented by < speciesGlyph> and are grouped in a <listOfSpeciesGlyphs> tag. In addition to the attributes from GraphicalObject, the speciesGlyph object has a species attribute which is the id of the corresponding species object in the model. The species attribute is optional to allow the program to specify species representations that do not have a direct correspondence in the model. This might be useful if some pathway has been collapsed, but is still treated by layout programs.
XML Schema representation:
<xsd:complexType name="SpeciesGlyph"> <xsd:complexContent> <xsd:extension base="sl2:GraphicalObject"> <xsd:attribute name="species" type="sl2:SId" use="optional"/> </xsd:extension> </xsd:complexContent> </xsd:complexType>
example:
.
.
.
<species id="ATP" compartment="compartment" initialAmount="0">
.
.
.
<sl2:listOfSpeciesGlyphs>
<sl2:speciesGlyph id="ATP_Glyph" species="ATP" x="295.0" y="123.0" w="16.0"/>
.
.
.
</sl2:listOfSpeciesGlyphs>
.
.
.
The reaction layout is now also a subclass of GraphicalObject, therefore it has a defualt bounding box attribute. This differs from the last versions of this document, where the reaction layout consisted of two pseudo nodes. Actually this is only a renaming of attributes to make it more consistent with compartment and species layout information. The two points given by the bounding box can still be interpreted as being the two pseudo nodes of a reaction. The reaction attribute specifies the id of the corresponding reaction in the model. Again, this reference is optional. The ReactionGlyph also holds a listOfSpeciesReferenceGlyphs (see below) which correspond to the layout representations of the reactants of the reaction in the model.
XML Schema representation:
<xsd:complexType name="ListOfSpeciesReferenceGlyphs">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="speciesReferenceGlyph" type="sl2:SpeciesReferenceGlyph"
minOccurs="1" maxOccurs="unbounded"/>
</xsd:sequence>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="ReactionGlyph">
<xsd:complexContent>
<xsd:extension base="sl2:GraphicalObject">
<xsd:sequence>
<xsd:element name="listOfSpeciesReferenceGlyphs"
type="sl2:ListOfSpeciesReferenceGlyphs"
minOccurs="1" maxOccurs="1"/>
</xsd:sequence>
<xsd:attribute name="reaction" type="sl2:SId" use="optional"/>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
example:
.
.
.
<reaction id="reaction_0" reversible="false">
<listOfReactants>
<speciesReference species="P_i" stoichiometry="1"/>
.
.
.
</reaction>
.
.
.
<sl2:listOfReactionGlyphs>
<sl2:reactionGlyph id="reaction_0_Glyph" reaction="reaction_0"
x1="119.0" y1="166.0" x2="120.0" y2="183.0"/>
.
.
.
</sl2:listOfReactionGlyphs>
.
.
.
The graphical connection between a speciesGlyph and a reactionGlyph (which would be an arrow or some curve in most cases) is represented by the speciesReferenceGlyph object. A listOfSpeciesReferenceGlyphs is contained in a reactionGlyph.
The speciesReferenceGlyph has a speciesGlyph attribute that contains the id of the graphical object that is to be connected to the reactionGlyph. This can be the id of a graphical representation of a species (as defined in the <listOfSpeciesGlyphs> ) or the id of a group (see below). The speciesReference attribute refers to a speciesReference in the model and is optional. Since species references in sbml level 1 as well as level 2 do not have ids, we chose to put a new tag called id which has an attribute called id that is of type sl2:SId into the annotation part of the corresponding SpeciesReference element. This tag has to be unique within the global namespace of the SBML model and can thus be used to reference a given species reference. The role attribute is used to specify how the species reference should be displayed. Allowed values are substrate, product, sidesubstrate, sideproduct, modifier, activator and inhibitor. This attribute is optional and should only be necessary if the optional speciesReference attribute is not given or if the respective information from the model needs to be overridden. The values substrate and product are used if the species reference is a main product or substrate in the reaction. sidesubstrate and sideproduct are used for stuff like ATP, NAD+, etc. that some renderers might choose to display as side reactants. activator and inhibitor are modifiers where their influence on the reaction is known and modifier is a more general term if the influence is unknown or changes during the course of the simulation. This list is probably not exhaustive and will be updated as needed. Future version of SBML may very well have an id for the SpeciesReference objects as well as some kind of role attribute. If this is the case, we will drop both attributes here since they are no longer necessary.
So far we have defined which graphical objects should be connected to the reaction glyph in which way. This is the minimum information that a render program with biochemical knowledge needs to render the reaction layout. For generality two alternative ways to specify more detailed layout information are provided. In most cases the relation of a species to a reaction will be graphically represented by a curve. In this case a curve tag that contains a listOfCurveSegments can be used. The listOfCurveSegments contains an arbitrary number of curve segments. For now we provide the definitions for two types of curve segments (LineSegment and CubicBezier) but leave it open if this should in future be restricted to only one type or even generalized to more different line types. All segment types, which is just CubicBezier so far, are derived from the LineSegment type. The type of the curve segment has to be specified with a xsi:type attribute in the curveSegment tag. The LineSegment just specifies the start and the end point of a line segment which should be a common feature of all types of curve segments, the CubicBezier additionally defines two control points needed to specify a cubic Bezier curve segment. If the curve attribute is specified this takes precedence over the bounding box defined by the attributes of the GraphicalObject from which SpeciesReferenceGlyph inherits.
XML Schema representation:
<xsd:complexType name="LineSegment">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:attribute name="x1" type="xsd:double"/>
<xsd:attribute name="y1" type="xsd:double"/>
<xsd:attribute name="z1" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="x2" type="xsd:double"/>
<xsd:attribute name="y2" type="xsd:double"/>
<xsd:attribute name="z2" type="xsd:double" use="optional" default="0.0"/>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="CubicBezier">
<xsd:complexContent>
<xsd:extension base="sl2:LineSegment">
<xsd:attribute name="x3" type="xsd:double"/>
<xsd:attribute name="y3" type="xsd:double"/>
<xsd:attribute name="z3" type="xsd:double" use="optional" default="0.0"/>
<xsd:attribute name="x4" type="xsd:double"/>
<xsd:attribute name="y4" type="xsd:double"/>
<xsd:attribute name="z4" type="xsd:double" use="optional" default="0.0"/>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="ListOfCurveSegments">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="curveSegment" type="sl2:LineSegment"
minOccurs="1" maxOccurs="unbounded"/>
</xsd:sequence>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="Curve">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="listOfCurceSegments" type="sl2:ListOfCurveSegments"/>
</xsd:sequence>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:simpleType name="RoleString">
<xsd:restriction base="xsd:string">
<xsd:enumeration value="substrate"/>
<xsd:enumeration value="product"/>
<xsd:enumeration value="sidesubstrate"/>
<xsd:enumeration value="sideproduct"/>
<xsd:enumeration value="modifier"/>
<xsd:enumeration value="activator"/>
<xsd:enumeration value="inhibitor)"/>
</xsd:restriction>
</xsd:simpleType>
<xsd:complexType name="SpeciesReferenceGlyph">
<xsd:complexContent>
<xsd:extension base="sl2:GraphicalObject">
<xsd:sequence>
<xsd:element name="curve" type="sl2:Curve" minOccurs="0" maxOccurs="1"/>
</xsd:sequence>
<xsd:attribute name="speciesGlyph" type="sl2:SId" use="optional"/>
<xsd:attribute name="speciesReference" type="sl2:SId" use="optional"/>
<xsd:attribute name="role" type="sl2:RoleString" use="optional"/>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
example:
.
.
.
<sl2:speciesGlyph id="P_iGlyph" species="P_i_s1" x="0.0" y="1.8"
w="1.0" h="1.0"/>
.
.
.
<sl2:reactionGlyph id="reaction_0_Glyph" reaction="reaction_0"
x1="2.3" y1="1.0" x2="3.0" y2="1.0">
<sl2:listOfSpeciesReferenceGlyphs>
<sl2:speciesReferenceGlyph id="SP1_Glyph" speciesGlyph="P_iGlyph"
speciesReference="P_i_sr1">
<sl2:curve>
<sl2:listOfCurveSegments>
<sl2:curveSegment xsi:type="sl2:CubicBezier" x1="0.0" y1="1.8"
x2="0.3" y2="0.8"
cx1="0.1" cy1="1.9" cx2="0.3" cy2="0.7"/>
<sl2:curveSegment xsi:type="sl2:LineSegment" x1="0.3" y1="0.8"
x2="2.3" y2="1.0" />
<sl2:/listOfCurveSegments>
<sl2:/curve>
</sl2:speciesReferenceGlyph>
.
.
.
</sl2:listOfSpeciesReferenceGlyphs>
</sl2:reactionGlyph>
.
.
XML Schema representation:
<xsd:complexType name="Component">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:attribute name="ref" type="sl2:SId"/>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
<xsd:complexType name="LayoutGroup">
<xsd:complexContent>
<xsd:extension base="sl2:SBase">
<xsd:sequence>
<xsd:element name="component" type="sl2:Component" minOccurs="2"
maxOccurs="unbounded"/>
</xsd:sequence>
<xsd:attribute name="id" type="sl2:SId"/>
</xsd:extension>
</xsd:complexContent>
</xsd:complexType>
We already started on the definition of the render part of this proposal. Until is has reached a certain level of maturity we will keep those two documents separate. Eventually though, they will end up in one common proposal document. This document is a work in progress and can be subject to changes any time. We are glad for any suggestions, corrections or hints to further improve these extensions. Hopefully with some help we could come up with a set of extensions that would suite the needs of many applications developed in this area. This specification was intended for the use with sbml level 2. With some slight changes it can also be used with sbml level 1 documents. Actually the only major changes that would have to be made is to change all references to SId to be references to SNames.
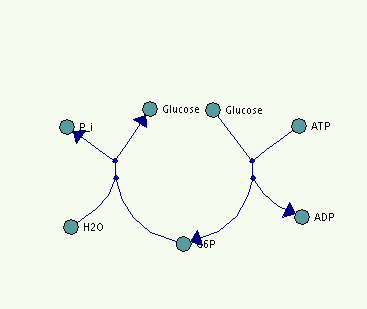
Last but not least, we include a small sample file to illustrate and complement the paragraphs above. Note that both the picture and the example code were manually modified. There does not exist an actual implementation of this latest version of our proposal yet. The model consists of two reactions. Which are the first reaction of glycolysis where glucose is converted to glucose-6-phosphate (G6P) and the reverse reaction of gluconeogenesis where glucose-6-phosphate is hydrolyzed to glucose. We did not include any coordinates in the third dimension, since we are only working in 2D space so far. As can be seen in the screenshot, the glucose SpeciesReference has two representational objects on screen whereas glucose-6-phosphate only has one. This difference is reflected in the file where the glucose species has two nodes in the listOfNodes whereas G6P only has one. This example show not all but only the main features of our proposal.
 |
<?xml version="1.0" encoding="UTF-8"?>
<sbml xmlns="http://www.sbml.org/sbml/level2" level="2" version="1"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:sl2="http://projects.eml.org/bcb/sbml/level2"
xsi:schemaLocation="http://projects.eml.org/bcb/sbml/level2
http://projects.eml.org/bcb/sbml/level2/layout2.xsd">
<model name="Untitled">
<listOfCompartments>
<compartment id="compartment" volume="1"/>
</listOfCompartments>
<listOfSpecies>
<species id="ATP" compartment="compartment" initialAmount="0"/>
<species id="P_i" compartment="compartment" initialAmount="0"/>
<species id="Glucose" compartment="compartment" initialAmount="0"/>
<species id="ADP" compartment="compartment" initialAmount="0"/>
<species id="H2O" compartment="compartment" initialAmount="0"/>
<species id="G6P" compartment="compartment" initialAmount="0"/>
</listOfSpecies>
<listOfReactions>
<reaction id="reaction_0" reversible="false">
<listOfReactants>
<speciesReference species="H2O" stoichiometry="1">
<annotation>
<sl2:id id="H2O"/>
</annotation>
</speciesReference>
<speciesReference species="G6P" stoichiometry="1">
<annotation>
<sl2:id id="G6P_1"/>
</annotation>
</speciesReference>
</listOfReactants>
<listOfProducts>
<speciesReference species="P_i" stoichiometry="1">
<annotation>
<sl2:id id="P_i_sr1"/>
</annotation>
</speciesReference>
<speciesReference species="Glucose" stoichiometry="1">
<annotation>
<sl2:id id="Glucose_1"/>
</annotation>
</speciesReference>
</listOfProducts>
</reaction>
<reaction id="reaction_1" reversible="false">
<listOfReactants>
<speciesReference species="ATP" stoichiometry="1">
<annotation>
<sl2:id id="ATP"/>
</annotation>
</speciesReference>
<speciesReference species="Glucose" stoichiometry="1">
<annotation>
<sl2:id id="Glucose_2"/>
</annotation>
</speciesReference>
</listOfReactants>
<listOfProducts>
<speciesReference species="ADP" stoichiometry="1">
<annotation>
<sl2:id id="ADP"/>
</annotation>
</speciesReference>
<speciesReference species="G6P" stoichiometry="1">
<annotation>
<sl2:id id="G6P_2"/>
</annotation>
</speciesReference>
</listOfProducts>
</reaction>
</listOfReactions>
</model>
<annotation>
<sl2:listOfLayouts xmlns:sl2="http://projects.eml.org/bcb/sbml/level2"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://projects.eml.org/bcb/sbml/level2\
http://projects.eml.org/bcb/sbml/level2/layout2.xsd">
<sl2:layout id="layout1" width="320" height="270">
<sl2:listOfCompartmentGlyphs>
<sl2:compartmentGlyph id="compGlyph" compartment="compartment"
x="10.0" y="10.0" w="60" h="50"/>
</sl2:listOfCompartmentGlyphs>
<sl2:listOfSpeciesGlyphs>
<sl2:speciesGlyph id="ATP_Glyph" species="ATP" x="295.0" y="123.0"
w="16.0" h="16.0"/>
<sl2:speciesGlyph id="P_iGlyph" species="P_i" x="63.0" y="124.0"
w="16.0" h="16.0"/>
<sl2:speciesGlyph id="Glucose_Glyph1" species="Glucose" x="146.0" y="106.0"
w="16.0" h="16.0"/>
<sl2:speciesGlyph id="Glucose_Glyph2" species="Glucose" x="209.0" y="107.0"
w="16.0" h="16.0"/>
<sl2:speciesGlyph id="ADP_Glyph" species="ADP" x="298.0" y="214.0"
w="16.0" h="16.0"/>
<sl2:speciesGlyph id="H2O_Glyph" species="H2O" x="67.0" y="224.0"
w="16.0" h="16.0"/>
<sl2:speciesGlyph id="G6P_Glyph" species="G6P" x="180.0" y="241.0"
w="15.0" h="16.0"/>
</sl2:listOfSpeciesGlyphs>
<sl2:listOfReactionGlyphs>
<sl2:reactionGlyph id="reaction_0_Glyph" reaction="reaction_0"
x="120.0" y="183.0" width="119.0" height="166.0">
<sl2:listOfSpeciesReferenceGlyphs>
<sl2:speciesReferenceGlyph id="SP1_Glyph" speciesGlyph="P_i_Glyph"
speciesReference="P_i_sr1"/>
<sl2:speciesReferenceGlyph id="SP2_Glyph" speciesGlyph="Glucose_Glyph1"
speciesReference="Glucose_1" role="substrate"/>
<sl2:speciesReferenceGlyph id="SP3_Glyph" speciesGlyph="H2O_Glyph"
speciesReference="H2O"/>
<sl2:speciesReferenceGlyph id="SP4_Glyph" speciesGlyph="G6P_Glyph"
speciesReference="G6P_1">
<sl2:curve>
<sl2:listOfCurveSegments>
<sl2:curveSegment xsi:type="sl2:CubicBezier" x1="180" y1="241"
x2="120" y2="183"
cx1="155" cy1="230" cx2="120" cy2="200"/>
<sl2:/listOfCurveSegments>
<sl2:/curve>
</sl2:speciesReferenceGlyph>
</sl2:listOfSpeciesReferenceGlyphs>
</sl2:reactionGlyph>
<sl2:reactionGlyph id="reaction_1_Glyph" reaction="reaction_1"
x1="256.0" y1="166.0" x2="257.0" y2="183.0">
<sl2:listOfSpeciesReferenceGlyphs>
<sl2:speciesReferenceGlyph id="SP5_Glyph" speciesGlyph="ATP_Glyph"
speciesReference="ATP"/>
<sl2:speciesReferenceGlyph id="SP6_Glyph" speciesGlyph="Glucose_Glyph2"
speciesReference="Glucose_2"/>
<sl2:speciesReferenceGlyph id="SP7_Glyph" speciesGlyph="ADP_Glyph"
speciesReference="ADP"/>
<sl2:speciesReferenceGlyph id="SP8_Glyph" speciesGlyph="G6P_Glyph"
speciesReference="G6P_2">
<sl2:curve>
<sl2:listOfCurveSegments>
<sl2:curveSegment xsi:type="sl2:CubicBezier" x1="257" y1="183"
x2="180" y2="241"
cx1="257" cy1="200" cx2="210" cy2="230"/>
<sl2:/listOfCurveSegments>
<sl2:/curve>
</sl2:speciesReferenceGlyph>
</sl2:listOfSpeciesReferenceGlyphs>
</sl2:reactionGlyph>
</sl2:listOfReactionGlyphs>
<sl2:listOfAdditionalGraphicalObjects>
<textLabel id="label1" x="100.0" y="85.0"
w="180.0" h="25.0">Reaction Layout</textLabel>
</sl2:listOfAdditionalGraphicalObjects>
</sl2:layout>
</sl2:listOfLayouts>
</annotation>
</sbml>
FIRSTNAME.LASTNAME@eml.villa-bosch.de
e.g.
ralph.gauges@eml.villa-bosch.de